Introduction to Operon Theory
Table of Contents
ToggleIn an operon theory, Operon is a functional unit of DNA found in prokaryotic organisms, such as bacteria. It consists of a set of genes that are transcribed together into a single mRNA molecule under the control of the Operator gene. This coordinated transcription allows for the regulation and expression of multiple genes simultaneously, often involved in related metabolic pathways or cellular functions.
Components of the Operon Model
The operon model comprises several key components that work together to regulate gene expression in prokaryotic organisms. These components include:
- Structural Genes: Structural genes are segments of DNA that encode the synthesis of specific proteins or enzymes. They are transcribed into messenger RNA (mRNA) during gene expression, ultimately leading to the production of functional proteins within the cell.
- Operator: The operator is a region of DNA located adjacent to the structural genes within the operon. It serves as a regulatory site where a repressor protein can bind. The binding of the repressor to the operator can block the transcription of the structural genes, thereby controlling their expression.
- Promoter: The promoter is another DNA sequence found upstream of the structural genes in the operon. It provides binding site for RNA polymerase, the enzyme responsible for initiating transcription. The promoter plays a crucial role in initiating the transcription process and determining the rate of gene expression.
- Regulatory Genes: Regulatory genes encode proteins that interact with the operator or promoter regions to regulate the expression of the structural genes within the operon. These regulatory proteins can act as activators or repressors, influencing the transcriptional activity of the operon in response to various environmental signals.
Together, these components form the basis of the operon model, facilitating the precise control of gene expression in prokaryotic organisms.

Functioning of Operon Model
The functioning of the operon model involves two main types of operons: inducible operons and repressible operons.
Inducible Operons:
Inducible operons are typically inactive but can be turned on, or induced, in response to specific environmental signals or substrates. When the inducer molecule is present, it interacts with a regulatory protein (such as a repressor), causing a conformational change that prevents the repressor from binding to the operator region. As a result, RNA polymerase can bind to the promoter and initiate transcription of the structural genes within the operon. An example of an inducible operon is the lac operon in Escherichia coli, which is activated in the presence of lactose when glucose is scarce.
Repressible Operons:
Repressible operons, on the other hand, are typically active but can be turned off, or repressed, in response to specific regulatory molecules. In this case, the regulatory molecule, often a corepressor, binds to the regulatory protein, altering its conformation so that it can bind to the operator region. This binding prevents RNA polymerase from transcribing the structural genes. An example of a repressible operon is the trp operon, which controls the synthesis of tryptophan biosynthetic enzymes in bacteria. When tryptophan levels are high, it acts as a corepressor, leading to the repression of the operon.
Examples of Operon Model
Lac Operon: Activation by Lactose
The lac operon is a well-studied genetic regulatory system found in bacteria, particularly Escherichia coli (E. coli). It consists of a set of genes involved in the metabolism of lactose, a sugar found in milk and other dairy products. The lac operon provides a classic example of an inducible operon, meaning it is activated in response to specific environmental signals.
Components of the Lac Operon:
- Structural Genes: The lac operon contains three structural genes: lacZ, lacY, and lacA.
- lacZ: Encodes beta-galactosidase, an enzyme that hydrolyzes lactose into glucose and galactose.
- lacY: Encodes lactose permease, a membrane protein that facilitates the entry of lactose into the bacterial cell.
- lacA: Encodes transacetylase, an enzyme involved in the detoxification of certain lactose derivatives.
- Operator and Promoter: The lac operon also includes an operator region and a promoter region.
- Operator: Located between the promoter and the structural genes, the operator is where the lac repressor protein binds to regulate gene expression.
- Promoter: The promoter is the DNA sequence where RNA polymerase binds to initiate transcription of the structural genes.
- Regulatory Gene: The lac operon is regulated by the lacI gene, which is situated upstream of the operon and encodes the lac repressor protein.
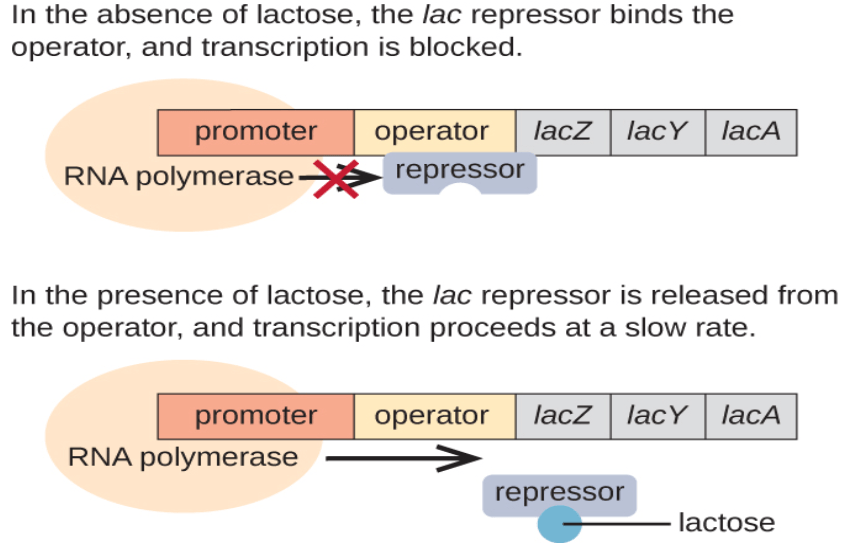
Regulation of the Lac Operon:
Under normal conditions, the lac repressor protein binds to the operator, blocking the binding of RNA polymerase to the promoter and preventing transcription of the structural genes. This occurs in the absence of lactose.
However, when lactose is present in the environment, it serves as an inducer molecule. Lactose is converted into allolactose, which binds to the lac repressor protein, causing it to undergo a conformational change. As a result, the repressor can no longer bind to the operator, allowing RNA polymerase to access the promoter and initiate transcription of the lac genes.
Significance of the Lac Operon:
The lac operon allows bacteria like E. coli to efficiently metabolize lactose when it is available in their environment. By tightly regulating the expression of the lac genes based on the presence or absence of lactose, bacteria can conserve energy by only producing the necessary enzymes when needed.
Overall, the lac operon serves as a classic model for understanding gene regulation and the mechanisms of inducible operons in bacteria. Its study has contributed significantly to our understanding of molecular biology and gene expression regulation.
Lac Operon: Activation by Catabolite Activator Protein (CAP)
The lac operon, found in bacteria like Escherichia coli (E. coli), is a classic example of gene regulation through positive control. It demonstrates how the presence of specific regulatory proteins can activate gene expression in response to environmental signals. One crucial regulatory protein involved in the activation of the lac operon is the catabolite activator protein (CAP).
Activation of the lac Operon by Catabolite Activator Protein (CAP):
- Background:
- The lac operon comprises three structural genes: lacZ, lacY, and lacA, involved in lactose metabolism.
- Under normal conditions, glucose is preferred substrate for the source of energy, and the lac operon is only weakly expressed even in the presence of lactose. This phenomenon is known as catabolite repression. Under depletion of glucose, bacteria have a mechanism that their genes to encode enzymes for using alternative sources of energy.
- When primary source glucose is consumed, the growth rate slows down, and then it switches from glucose to another substrate like lactose with the help of an enzyme called Enzyme IIA (EIIA). At a slow growth rate, cells produce less ATP for catabolism and EIIA becomes phosphorylated. And Phosphorylated EIIA activates adenylyl cyclase (an enzyme that converts some of the remaining ATP to cyclic AMP). Hence cAMP level begins to rise. cAMP is involved in glucose and energy metabolism.
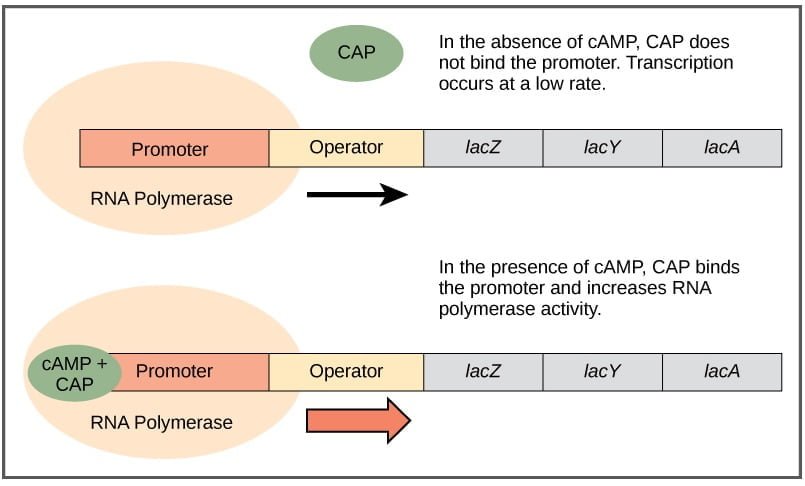
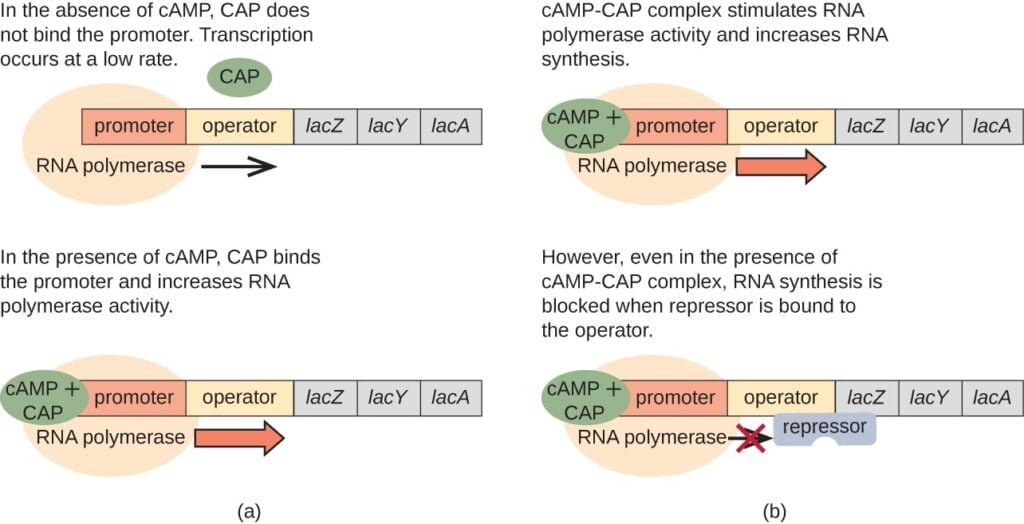
- Role of Catabolite Activator Protein (CAP):
- Catabolite activator protein also known as the cyclic AMP (cAMP) receptor protein, plays a crucial role in activating the lac operon.
- CAP is activated when levels of cyclic AMP (cAMP) increase in the cell, usually when glucose levels are low.
- Mechanism of Activation:
- When glucose levels are low, the concentration of cAMP increases in the cell.
- cAMP binds to CAP, forming a cAMP-CAP complex.
- The cAMP-CAP complex then binds to a specific DNA sequence called the CAP-binding site, located upstream of the lac operon promoter.
- Enhancement of Transcription:
- Binding of the cAMP-CAP complex to the CAP-binding site enhances the affinity of RNA polymerase for the lac operon promoter.
- This enhanced affinity promotes the binding of RNA polymerase to the promoter and facilitates the initiation of transcription of the lac genes.
- Increased Expression of Lac Genes:
- With the lac operon effectively activated by the cAMP-CAP complex, transcription of the lacZ, lacY, and lacA genes increases.
- The lacZ gene encodes β-galactosidase, which catalyzes the hydrolysis of lactose into glucose and galactose.
- The lacY gene encodes lactose permease, which facilitates the entry of lactose into the bacterial cell.
- The lacA gene encodes transacetylase, which is involved in the metabolism of lactose derivatives.
Conclusion: The activation of the lac operon by catabolite activator protein (CAP) demonstrates how bacteria regulate gene expression in response to changes in nutrient availability. By activating the lac operon in the absence of glucose and in the presence of lactose, bacteria like E. coli can efficiently utilize alternative carbon sources for energy production. This regulatory mechanism ensures that cells adapt to their environment and optimize their metabolic activities for survival.
The trp Operon : A Repressible Operon
The trp operon is another important genetic regulatory system found in bacteria, including Escherichia coli (E. coli). It is responsible for regulating the synthesis of tryptophan, an essential amino acid required for protein synthesis and various cellular processes. The trp operon exemplifies a repressible operon, meaning it is normally active but can be inhibited or repressed under specific conditions.
Components of the Trp Operon:
- Structural Genes: The trp operon consists of several genes involved in the biosynthesis of tryptophan.
- trpE, trpD, trpC, trpB, trpA: These structural genes encode enzymes involved in the different steps of tryptophan biosynthesis.
- Operator and Promoter: Similar to the lac operon, the trp operon contains an operator region and a promoter region.
- Operator: The operator is where the trp repressor protein binds to regulate gene expression.
- Promoter: The promoter is where RNA polymerase binds to initiate transcription of the structural genes.
- Regulatory Gene: The trp operon is regulated by the trpR gene, which codes for the trp repressor protein.
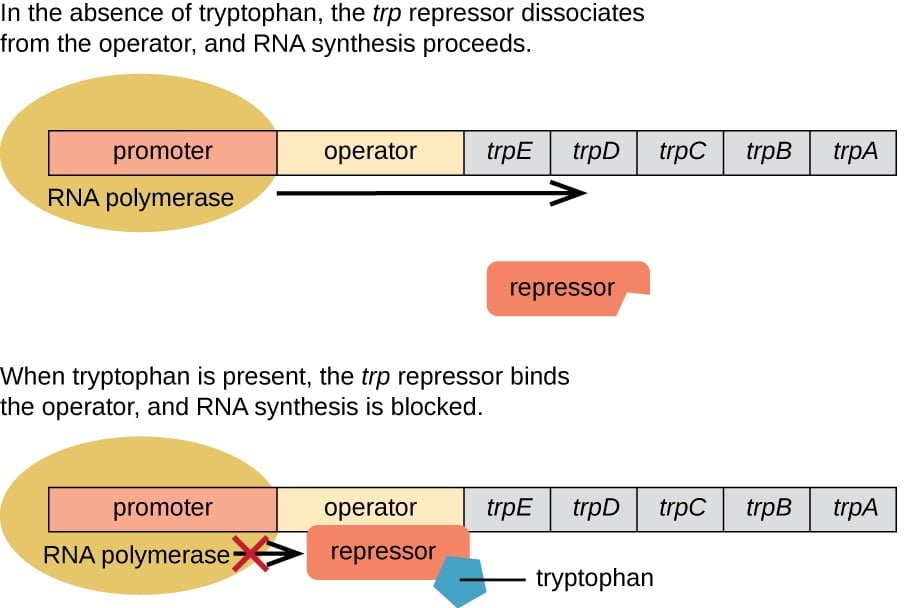
Regulation of the Trp Operon:
Under normal conditions, when the concentration of tryptophan in the cell is sufficient, tryptophan molecules bind to the trp repressor protein, causing it to undergo a conformational change. The activated repressor then binds to the operator, effectively blocking RNA polymerase from transcribing the structural genes of the trp operon. This regulatory mechanism prevents the unnecessary synthesis of tryptophan when it is already abundant in the cellular environment.
However, when the concentration of tryptophan decreases, there is less tryptophan available to bind to the repressor protein. As a result, the repressor remains inactive and cannot bind to the operator. This allows RNA polymerase to access the promoter and initiate transcription of the structural genes, leading to the synthesis of tryptophan.
Significance of the Trp Operon:
The trp operon enables bacteria to regulate the synthesis of tryptophan in response to changes in the availability of this essential amino acid. By tightly controlling the expression of the trp genes, bacteria can efficiently utilize cellular resources and adapt to varying environmental conditions.
In summary, the trp operon represents a crucial mechanism of gene regulation in bacteria, illustrating how cells adjust their metabolic activities in response to the levels of specific metabolites.
Significance of the Operon Model in Gene Expression:
The significance of the operon model in gene expression includes two key aspects: the control of gene expression and adaptation to environmental changes.
- Control of Gene Expression:
With the help of operon theory, regulation and expression of genes in prokaryotic organisms can be understood. By coordinating the transcription of multiple genes involved in related metabolic pathways or cellular functions, operons allow for precise control of gene expression.
Through mechanisms such as positive and negative regulation, operons enable cells to modulate the synthesis of specific proteins in response to internal and external signals. This regulation ensures that genes are expressed at the right time and in the right amounts to meet the metabolic needs of the cell.
- Adaptation to Environmental Changes:
Operons play an important role in organisms to adapt to changing environmental conditions. By regulating gene expression in response to fluctuations in nutrient availability, temperature, pH, and other environmental factors, cells can optimize their metabolic activities and enhance their survival chances.
For example, inducible operons allow cells to activate specific metabolic pathways only when the corresponding substrates are present in the environment. This adaptive response ensures efficient utilization of available resources and minimizes energy expenditure on unnecessary metabolic processes.
Similarly, repressible operons enable cells to downregulate the synthesis of certain proteins when their substrates are abundant, preventing the wasteful use of resources. This regulatory mechanism helps maintain cellular homeostasis and ensures the efficient allocation of metabolic resources under varying environmental conditions.
Historical Development of Operon Model
The historical development of the operon model is most important in molecular biology. François Jacob and Jacques Monod played fabulous roles in providing the principles underlying gene regulation in bacteria.
Discoveries Leading to the Operon Model:
- Early Observations: In the mid-20th century, researchers observed peculiar patterns of gene expression in bacteria. They noted that certain genes appeared to be coordinately regulated and expressed together, suggesting the presence of regulatory mechanisms governing gene activity.
- The Lac Operon: In the 1950s and 1960s, François Jacob and Jacques Monod conducted groundbreaking research on the lac operon in Escherichia coli. They observed that genes involved in lactose metabolism were under coordinated control, with their expression being induced in the presence of lactose.
- Regulatory Mechanisms: Through a series of elegant experiments, Jacob and Monod uncovered the regulatory mechanisms governing the lac operon. They proposed the existence of an operon as a genetic switch that could be turned on or off based on the availability of lactose and glucose in the bacterial environment.
Contributions of Jacob and Monod:
- Conceptual Framework: Jacob and Monod developed the operon model, which provided a conceptual framework for understanding gene regulation in bacteria. They proposed that the lac operon consisted of structural genes, a promoter region for RNA polymerase binding, and an operator site where regulatory proteins could interact.
- Inducer-Repressor System: Their studies revealed the role of inducers, such as lactose, in activating the lac operon by alleviating the inhibitory effects of a repressor protein. They proposed that the repressor protein could bind to the operator site, blocking transcription of the structural genes in the absence of lactose.
- Genetic Switch: Jacob and Monod introduced the concept of a genetic switch, whereby the lac operon could be toggled between active and inactive states depending on the presence of inducers and repressors. This model provided a mechanistic explanation for the regulation of gene expression in response to environmental signals.
Modern Applications of Operon Model
The operon model, initially discovered by François Jacob and Jacques Monod in the 1960s, has found modern applications in various fields, particularly genetic engineering and biotechnology. Let’s delve into how the operon model is used in these areas:
- Genetic Engineering:
Understanding Gene Regulation:
- The operon model provides insights into how genes are regulated and expressed in organisms.
- By understanding the regulatory mechanisms, genetic engineers can manipulate gene expression for various purposes.
Inducible Promoters:
- Inducible promoters derived from operons are often used in genetic engineering.
- These promoters can be activated by specific inducers, allowing precise control over gene expression in engineered organisms.
Gene Expression Systems:
- The lac operon, with its inducible promoter, is commonly used in gene expression systems.
- Researchers can insert genes of interest downstream of the lac promoter and control their expression by adding or removing inducers.
- Biotechnology:
Production of Recombinant Proteins:
- The operon model is instrumental in producing recombinant proteins for various applications.
- Recombinant DNA technology allows the insertion of genes encoding desired proteins into bacterial cells under the control of inducible promoters.
- By inducing expression using specific inducers, large quantities of recombinant proteins can be produced.
Metabolic Engineering:
- Metabolic pathways can be engineered using the operon model to enhance the production of desired compounds.
- By manipulating the expression of genes involved in metabolic pathways, biotechnologists can optimize the production of biofuels, pharmaceuticals, and other valuable chemicals.
Synthetic Biology:
- Synthetic biology aims to design and construct novel biological systems for specific purposes.
- The operon model serves as a framework for designing synthetic gene circuits and regulatory networks.
- Synthetic biologists can engineer cells with custom-designed operons to perform complex tasks, such as sensing environmental signals or producing specific molecules.
FAQs :
- What is an operon?
- An operon is a genetic regulatory system found in prokaryotic organisms, consisting of a cluster of genes under the control of a single promoter and regulatory elements.
- What is the role of an operator in an operon?
- The operator is a DNA sequence located near the promoter region of the operon. It acts as a binding site for regulatory proteins, such as repressors, which can modulate the transcription of genes within the operon.
- What are inducible operons?
- Inducible operons are operons whose transcription is normally turned off but can be induced, or activated, in response to specific environmental signals or substrates.
- Can you provide an example of an inducible operon?
- The lac operon in Escherichia coli is a classic example of an inducible operon. It is activated in the presence of lactose, which serves as an inducer molecule.
- What are repressible operons?
- Repressible operons are operons that are typically active but can be inhibited, or repressed, in response to specific regulatory molecules or signals.
- Could you give an example of a repressible operon?
- The trp operon in E. coli is an example of a repressible operon. It is repressed in the presence of tryptophan, which acts as a corepressor molecule.
- How do inducible and repressible operons differ in their regulation?
- Inducible operons are activated in response to the presence of an inducer molecule, while repressible operons are inhibited in response to the presence of a corepressor molecule.
- What is the significance of the operon model in molecular biology?
- The operon model provides a framework for understanding how genes are regulated and expressed in prokaryotic organisms. It offers insights into the mechanisms of gene regulation and has applications in genetic engineering, biotechnology, and synthetic biology.
- How do regulatory proteins control gene expression in operons?
- Regulatory proteins, such as activators and repressors, bind to specific DNA sequences within operons and modulate the accessibility of RNA polymerase to the promoter region, thereby regulating transcription initiation.
- What are some current research areas related to operon theory?
- Current research areas include investigating the role of operons in microbial adaptation, exploring novel regulatory mechanisms within operons, and applying operon-based gene expression systems in synthetic biology and biotechnology.

